How To Add A Methyl Group To A Carbon Chain
In the chemic sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or grouping) by a methyl group. Methylation is a class of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil scientific discipline, and the biological sciences.
In biological systems, methylation is catalyzed by enzymes; such methylation tin exist involved in modification of heavy metals, regulation of gene expression, regulation of poly peptide function, and RNA processing. In vitro methylation of tissue samples is too one method for reducing certain histological staining artifacts. The counterpart of methylation is called demethylation.
In biology [edit]
In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate factor expression, RNA processing and poly peptide part. It has been recognized equally a key procedure underlying epigenetics.
Methanogenesis [edit]
Methanogenesis, the process that generates methane from COii, involves a series of methylation reactions. These reactions are effected past a set of enzymes harbored by a family of anaerobic microbes.[ane]
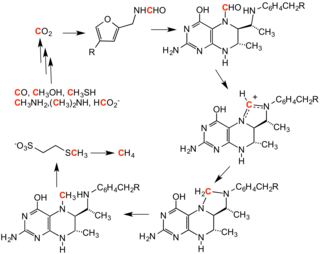
Cycle for methanogenesis, showing intermediates.
In opposite methanogenesis, marsh gas serves as the methylating agent.[ citation needed ]
O-methyltransferases [edit]
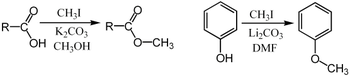
A wide variety of phenols undergo O-methylation to give anisole derivatives. This process, catalyzed by enzymes such as caffeoyl-CoA O-methyltransferase, is a key reaction in the biosynthesis of lignols, percursors to lignin, a major structural component of plants.
Plants produce flavonoids and isoflavones with methylations on hydroxyl groups, i.east. methoxy bonds. This 5-O-methylation affects the flavonoid´southward water solubility. Examples are 5-O-methylgenistein, 5-O-methylmyricetin or five-O-methylquercetin, besides known as azaleatin.
Proteins [edit]
Together with ubiquitin and phosphorylation, methylation is a major biochemical process for modifying protein role. The well-nigh prevalent protein methylations bear on arginine and lysine balance of specific histones. Otherwise histidine, glutamate, asparagine, cysteine are susceptible to methylation. Some of these products include S-methylcysteine, two isomers of North-methylhistidine, and 2 isomers of Northward-methylarginine.[2]
Methionine synthase [edit]

Methionine synthase regenerates methionine (Met) from homocysteine (Hcy). The overall reaction transforms five-methyltetrahydrofolate (Nv-MeTHF) into tetrahydrofolate (THF) while transferring a methyl group to Hcy to form Met. Methionine Synthases tin be cobalamin-dependent and cobalamin-independent: Plants take both, animals depend on the methylcobalamin-dependent form.
In methylcobalamin-dependent forms of the enzyme, the reaction proceeds by ii steps in a ping-pong reaction. The enzyme is initially primed into a reactive state by the transfer of a methyl group from N5-MeTHF to Co(I) in enzyme-bound cobalamin (Cob), forming methyl-cobalamin(Me-Cob) that now contains Me-Co(III) and activating the enzyme. So, a Hcy that has coordinated to an enzyme-bound zinc to form a reactive thiolate reacts with the Me-Cob. The activated methyl grouping is transferred from Me-Cob to the Hcy thiolate, which regenerates Co(I) in Cob, and Met is released from the enzyme.[3]
Heavy metals: arsenic, mercury, cadmium [edit]
Biomethylation is the pathway for converting some heavy elements into more mobile or more lethal derivatives that can enter the food concatenation. The biomethylation of arsenic compounds starts with the formation of methanearsonates. Thus, trivalent inorganic arsenic compounds are methylated to requite methanearsonate. Southward-adenosylmethionine is the methyl donor. The methanearsonates are the precursors to dimethylarsonates, again by the cycle of reduction (to methylarsonous acrid) followed by a second methylation.[4] Related pathways utilize to the biosynthesis of methylmercury.
Epigenetic methylation [edit]
DNA/RNA methylation [edit]
DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sites–that is, where a cytosine is direct followed by a guanine in the Deoxyribonucleic acid sequence). This methylation results in the conversion of the cytosine to five-methylcytosine. The formation of Me-CpG is catalyzed by the enzyme DNA methyltransferase. In mammals, DNA methylation is mutual in body cells,[five] and methylation of CpG sites seems to be the default.[6] [vii] Human Deoxyribonucleic acid has almost lxxx–xc% of CpG sites methylated, merely there are certain areas, known every bit CpG islands, that are CG-rich (loftier cytosine and guanine content, made up of about 65% CG residues), wherein none is methylated. These are associated with the promoters of 56% of mammalian genes, including all ubiquitously expressed genes. Ane to two percent of the human genome are CpG clusters, and at that place is an inverse relationship between CpG methylation and transcriptional activity. Methylation contributing to epigenetic inheritance can occur through either DNA methylation or protein methylation. Improper methylations of human genes can lead to disease development,[eight] [nine] including cancer.[ten] [xi] Similarly, RNA methylation occurs in dissimilar RNA species viz. tRNA, rRNA, mRNA, tmRNA, snRNA, snoRNA, miRNA, and viral RNA. Dissimilar catalytic strategies are employed for RNA methylation past a variety of RNA-methyltransferases. RNA methylation is idea to have existed before DNA methylation in the early forms of life evolving on earth.[12]
N6-methyladenosine (m6A) is the near common and abundant methylation modification in RNA molecules (mRNA) present in eukaryotes. 5-methylcytosine (five-mC) as well unremarkably occurs in various RNA molecules. Contempo information strongly suggest that m6A and 5-mC RNA methylation affects the regulation of various biological processes such equally RNA stability and mRNA translation,[13] and that abnormal RNA methylation contributes to etiology of human diseases.[fourteen]
Protein methylation [edit]
Protein methylation typically takes place on arginine or lysine amino acrid residues in the poly peptide sequence.[15] Arginine can exist methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylarginine) or one on both nitrogens (symmetric dimethylarginine), by protein arginine methyltransferases (PRMTs). Lysine can be methylated one time, twice, or three times by lysine methyltransferases. Protein methylation has been almost studied in the histones. The transfer of methyl groups from S-adenosyl methionine to histones is catalyzed by enzymes known as histone methyltransferases. Histones that are methylated on certain residues tin can act epigenetically to repress or activate gene expression.[xvi] [17] Protein methylation is one blazon of post-translational modification.
Evolution [edit]
Methyl metabolism is very ancient and can be found in all organisms on earth, from bacteria to humans, indicating the importance of methyl metabolism for physiology.[eighteen] Indeed, pharmacological inhibition of global methylation in species ranging from human, mouse, fish, fly, round worm, institute, algae and cyanobacteria causes the same effects on their biological rhythms, demonstrating conserved physiological roles of methylation during evolution.[19]
In chemistry [edit]
The term methylation in organic chemistry refers to the alkylation process used to describe the delivery of a CH3 grouping.[20]
Electrophilic methylation [edit]
Methylations are unremarkably performed using electrophilic methyl sources such as iodomethane,[21] dimethyl sulfate,[22] [23] dimethyl carbonate,[24] or tetramethylammonium chloride.[25] Less mutual but more powerful (and more dangerous) methylating reagents include methyl triflate,[26] diazomethane,[27] and methyl fluorosulfonate (magic methyl). These reagents all react via SouthwardNorthward2 nucleophilic substitutions. For case, a carboxylate may be methylated on oxygen to give a methyl ester; an alkoxide salt RO− may be likewise methylated to give an ether, ROCH3; or a ketone enolate may be methylated on carbon to produce a new ketone.
The Purdie methylation is a specific for the methylation at oxygen of carbohydrates using iodomethane and silverish oxide.[28]
Eschweiler–Clarke methylation [edit]
The Eschweiler–Clarke reaction is a method for methylation of amines.[29] This method avoids the risk of quaternization, which occurs when amines are methylated with methyl halides.

Diazomethane and trimethylsilyldiazomethane [edit]
Diazomethane and the safer counterpart trimethylsilyldiazomethane methylate carboxylic acids, phenols, and even alcohols:
- RCO2H + tmsCHN2 + CHiiiOH → RCO2CH3 + CHiiiOtms + N2
The method offers the advantage that the side products are hands removed from the product mixture.[30]
Nucleophilic methylation [edit]
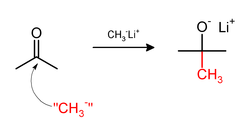
Methylation sometimes involve use of nucleophilic methyl reagents. Strongly nucleophilic methylating agents include methyllithium (CH3Li)[31] or Grignard reagents such every bit methylmagnesium bromide (CHiiiMgX).[32] For example, CHthreeLi will add methyl groups to the carbonyl (C=O) of ketones and aldehyde.:
Milder methylating agents include tetramethyltin, dimethylzinc, and trimethylaluminium.[33]
Run into besides [edit]
Biological science topics [edit]
- Bisulfite sequencing – the biochemical method used to determine the presence or absenteeism of methyl groups on a Dna sequence
- MethDB Deoxyribonucleic acid Methylation Database
- Microscale thermophoresis – a biophysical method to make up one's mind the methylisation state of DNA[34]
Organic chemical science topics [edit]
- Alkylation
- Methoxy
- Titanium–zinc methylenation
- Petasis reagent
- Nysted reagent
- Wittig reaction
- Tebbe's reagent
References [edit]
- ^ Thauer, R. K., "Biochemistry of Methanogenesis: a Tribute to Marjory Stephenson", Microbiology, 1998, book 144, pages 2377-2406.
- ^ Clarke, Steven G. (2018). "The ribosome: A hot spot for the identification of new types of protein methyltransferases". Journal of Biological Chemical science. 293 (27): 10438–10446. doi:10.1074/jbc.AW118.003235. PMC6036201. PMID 29743234.
- ^ Matthews, R. G.; Smith, A. E.; Zhou, Z. South.; Taurog, R. East.; Bandarian, V.; Evans, J. C.; Ludwig, 1000. (2003). "Cobalamin-Dependent and Cobalamin-Independent Methionine Synthases: Are There Two Solutions to the Same Chemical Problem?". Helvetica Chimica Acta. 86 (12): 3939–3954. doi:x.1002/hlca.200390329.
- ^ Styblo, M.; Del Razo, L. M.; Vega, L.; Germolec, D. R.; LeCluyse, E. L.; Hamilton, Grand. A.; Reed, W.; Wang, C.; Cullen, W. R.; Thomas, D. J. (2000). "Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and homo cells". Archives of Toxicology. 74 (half-dozen): 289–299. doi:10.1007/s002040000134. PMID 11005674. S2CID 1025140.
{{cite periodical}}: CS1 maint: uses authors parameter (link) - ^ Tost J (2010). "DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker". Mol Biotechnol. 44 (1): 71–81. doi:10.1007/s12033-009-9216-2. PMID 19842073. S2CID 20307488.
- ^ Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee Fifty, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (November 2009). "Human DNA methylomes at base resolution show widespread epigenomic differences". Nature. 462 (7271): 315–22. Bibcode:2009Natur.462..315L. doi:10.1038/nature08514. PMC2857523. PMID 19829295.
- ^ Stadler MB, Murr R, Burger 50, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schübeler D (December 2011). "DNA-binding factors shape the mouse methylome at distal regulatory regions". Nature. 480 (7378): 490–v. doi:10.1038/nature11086. PMID 22170606.
- ^ Rotondo JC, Selvatici R, Di Domenico M, Marci R, Vesce F, Tognon M, Martini F (September 2013). "Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males". Epigenetics. 8 (9): 990–vii. doi:10.4161/epi.25798. PMC3883776. PMID 23975186.
- ^ Rotondo JC, Bosi S, Bazzan E, Di Domenico G, De Mattei Chiliad, Selvatici R, Patella A, Marci R, Tognon M, Martini F (December 2012). "Methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples of infertile couples correlates with recurrent spontaneous abortion". Human Reproduction. 27 (12): 3632–viii. doi:10.1093/humrep/des319. PMID 23010533.
- ^ Rotondo JC, Borghi A, Selvatici R, Magri E, Bianchini Eastward, Montinari E, Corazza M, Virgili A, Tognon G, Martini F (2016). "Hypermethylation-Induced Inactivation of the IRF6 Factor equally a Possible Early Event in Progression of Vulvar Squamous Cell Carcinoma Associated With Lichen Sclerosus". JAMA Dermatology. 152 (eight): 928–33. doi:ten.1001/jamadermatol.2016.1336. PMID 27223861.
- ^ Rotondo JC, Borghi A, Selvatici R, Mazzoni E, Bononi I, Corazza 1000, Kussini J, Montinari East, Gafà R, Tognon M, Martini F (2018). "Association of Retinoic Acid Receptor β Gene With Onset and Progression of Lichen Sclerosus-Associated Vulvar Squamous Prison cell Carcinoma". JAMA Dermatology. 154 (vii): 819–823. doi:10.1001/jamadermatol.2018.1373. PMC6128494. PMID 29898214.
- ^ Rana, Ajay K.; Ankri, Serge (1 January 2016). "Reviving the RNA World: An Insight into the Advent of RNA Methyltransferases". Front Genet. 7: 99. doi:ten.3389/fgene.2016.00099. PMC4893491. PMID 27375676.
- ^ Choi, Junhong; Ieong, Ka-Weng; Demirci, Hasan; Chen, Jin; Petrov, Alexey; Prabhakar, Arjun; O'Leary, Seán E.; Dominissini, Dan; Rechavi, Gideon (February 2016). "N6-methyladenosine in mRNA disrupts tRNA choice and translation-elongation dynamics". Nature Structural & Molecular Biological science. 23 (2): 110–115. doi:10.1038/nsmb.3148. ISSN 1545-9993. PMC4826618. PMID 26751643.
- ^ Stewart, Kendal (15 September 2017). "Methylation (MTHFR) Testing & Folate Deficiency". Archived from the original on 12 October 2017. Retrieved 11 Oct 2017.
- ^ Walsh, Christopher (2006). "Affiliate 5 – Protein Methylation" (PDF). Posttranslational modification of proteins: expanding nature'due south inventory. Roberts and Co. Publishers. ISBN978-0-9747077-three-0. [ permanent expressionless link ]
- ^ Grewal, Due south. I.; Rice, J. C. (2004). "Regulation of heterochromatin by histone methylation and small-scale RNAs". Current Stance in Jail cell Biology. 16 (3): 230–238. doi:10.1016/j.ceb.2004.04.002. PMID 15145346.
- ^ Nakayama, J. -I.; Rice, J. C.; Strahl, B. D.; Allis, C. D.; Grewal, S. I. (2001). "Role of Histone H3 Lysine 9 Methylation in Epigenetic Control of Heterochromatin Assembly". Scientific discipline. 292 (5514): 110–113. Bibcode:2001Sci...292..110N. doi:10.1126/science.1060118. PMID 11283354. S2CID 16975534.
- ^ Kozbial, P.Z.; Mushegian, A.R. (2005). "Natural history of S-adenosylmethionine-bounden proteins". BMC Struct Biol. five (19): 19. doi:10.1186/1472-6807-v-xix. PMC1282579. PMID 16225687.
{{cite periodical}}: CS1 maint: uses authors parameter (link) - ^ Fustin, J.K.; Ye, S., Rakers, C.; Kaneko, K.; Fukumoto, K.; Yamano, Grand.; Versteven, G.; Grünewald, E.; Cargill, S.J.; Tamai, T.One thousand.; Xu, Y.; Jabbur, One thousand.L.; Kojima, R.; Lamberti, M.L.; Yoshioka-Kobayashi, K.; Whitmore, D.; Tammam, S.; Howell, P.L.; Kageyama, R.; Matsuo, T.; Stanewsky, R.; Golombek, D.A.; Johnson, C.H.; Kakeya, H.; van Ooijen, K.; Okamura, H. (2020). "Methylation deficiency disrupts biological rhythms from bacteria to humans". Communications Biological science. 3 (211): 211. doi:ten.1038/s42003-020-0942-0. PMC7203018. PMID 32376902.
{{cite periodical}}: CS1 maint: uses authors parameter (link) - ^ March, Jerry; Smith, Michael W (2001). March's advanced organic chemical science: reactions, mechanisms, and structure. New York: Wiley. ISBN978-0-471-58589-3.
- ^ Vyas, G. N.; Shah, N. M. (1951). "Quninacetophenone monomethyl ether". Organic Syntheses. 31: 90. doi:10.15227/orgsyn.031.0090.
- ^ Hiers, Chiliad. South. (1929). "Anisole". Organic Syntheses. 9: 12. doi:10.15227/orgsyn.009.0012.
- ^ Icke, Roland N.; Redemann, Ernst; Wisegarver, Burnett B.; Alles, Gordon A. (1949). "one thousand-Methoxybenzaldehyde". Organic Syntheses. 29: 63. doi:10.15227/orgsyn.029.0063.
- ^ Tundo, Pietro; Selva, Maurizio; Bomben, Andrea (1999). "Mono-C-methylathion of arylacetonitriles and methyl arylacetates by dimethyl carbonate: a general method for the synthesis of pure ii-arylpropionic acids. 2-Phenylpropionic acid". Organic Syntheses. 76: 169. doi:10.15227/orgsyn.076.0169.
- ^ Nenad, Maraš; Polanc, Slovenko; Kočevar, Marijan (2008). "Microwave-assisted methylation of phenols with tetramethylammonium chloride in the presence of 1000twoCO3 or Cs2CO3". Tetrahedron. 64 (51): 11618–11624. doi:10.1016/j.tet.2008.ten.024.
- ^ Poon, Kevin W. C.; Albiniak, Philip A.; Dudley, Gregory B. (2007). "Protection of alcohols using 2-benzyloxy-1-methylpyridinium trifluoromethanesulfanonate: Methyl (R)-(-)-three-benzyloxy-2-methyl propanoate". Organic Syntheses. 84: 295. doi:ten.15227/orgsyn.084.0295.
- ^ Neeman, M.; Johnson, William S. (1961). "Cholestanyl methyl ether". Organic Syntheses. 41: 9. doi:10.15227/orgsyn.041.0009.
- ^ Purdie, T.; Irvine, J. C. (1903). "C.?The alkylation of sugars". Journal of the Chemical Club, Transactions. 83: 1021–1037. doi:x.1039/CT9038301021.
- ^ Icke, Roland N.; Wisegarver, Burnett B.; Alles, Gordon A. (1945). "β-Phenylethyldimethylamine". Organic Syntheses. 25: 89. doi:10.15227/orgsyn.025.0089.
- ^ Shioiri, Takayuki; Aoyama, Toyohiko; Snowden, Timothy (2001). "Trimethylsilyldiazomethane". Encyclopedia of Reagents for Organic Synthesis. due east-EROS Encyclopedia of Reagents for Organic Synthesis. doi:10.1002/047084289X.rt298.pub2. ISBN978-0471936237.
{{cite encyclopedia}}: CS1 maint: uses authors parameter (link) - ^ Lipsky, Sharon D.; Hall, Stan Due south. (1976). "Aromatic Hydrocarbons from aromatic ketones and aldehydes: ane,1-Diphenylethane". Organic Syntheses. 55: 7. doi:10.15227/orgsyn.055.0007.
- ^ Grummitt, Oliver; Becker, Ernest I. (1950). "trans-ane-Phenyl-ane,3-butadiene". Organic Syntheses. 30: 75. doi:ten.15227/orgsyn.030.0075.
- ^ Negishi, Ei-ichi; Matsushita, Hajime (1984). "Palladium-Catalyzed Synthesis of i,iv-Dienes by Allylation of Alkenyalane: α-Farnesene". Organic Syntheses. 62: 31. doi:ten.15227/orgsyn.062.0031.
- ^ Wienken CJ, Baaske P, Duhr S, Braun D (2011). "Thermophoretic melting curves quantify the conformation and stability of RNA and Dna". Nucleic Acids Enquiry. 39 (8): e52. doi:10.1093/nar/gkr035. PMC3082908. PMID 21297115.
External links [edit]
| | Look upwards methylation in Wiktionary, the costless dictionary. |
- deltaMasses Detection of Methylations after Mass Spectrometry
How To Add A Methyl Group To A Carbon Chain,
Source: https://en.wikipedia.org/wiki/Methylation
Posted by: wardposs1950.blogspot.com

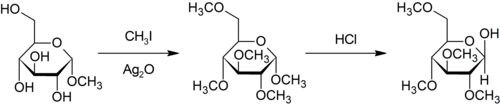

0 Response to "How To Add A Methyl Group To A Carbon Chain"
Post a Comment